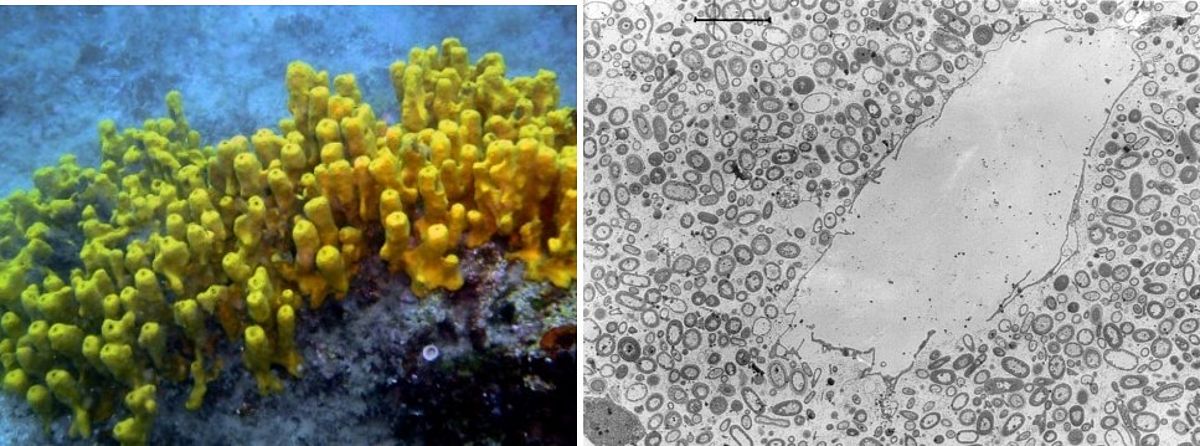
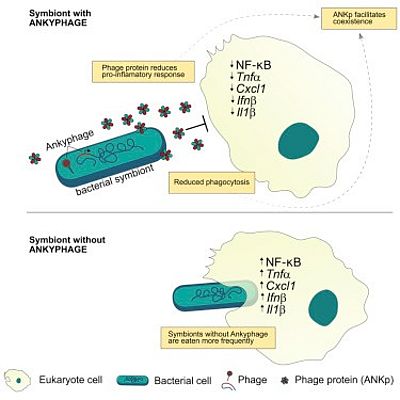
While recent studies have revealed vast diversity in the virosphere, the new frontier is to understand how phages modulate host-microbe interactions. We combine viral metagenomics with functional assays to investigate the interplay between phages, bacterial symbionts and marine sponges. Although sponges process massive amounts of water through their constantly operating filter apparatus, we have recently shown that they harbor species specific and even individually unique viral signatures distinct from other environments. We have further discovered an ankyrin protein (ANKp), encoded on a symbiont phage that, upon heterologous expression, modulates the eukaryotic immune response against bacteria as confirmed in macrophage infection assays. We hypothesize that these symbiont phage proteins facilitate co-existence in the multispecies interplay between phages, microbial symbionts and sponges in a manner that may extend to other host-microbe associations. Interestingly, these “Ankyphages” appear to be widely distributed in other host-associated contexts including human.
Researchers
Dr. Kristina Bayer, Dr. Martin T. Jahn, Dr. Laura Rix, Dr. Beate Slaby, Leon Steiner
Publications
Maldonado M, Bayer K, López-Acosta M (2024) Nitrogen and Phosphorus Cycling Through Marine Sponges: Physiology, Cytology, Genomics, and Ecological Implications. Frontiers in Invertebrate Physiology: A Collection of Reviews Apple Academic Press. eBook ISBN 9781003403319
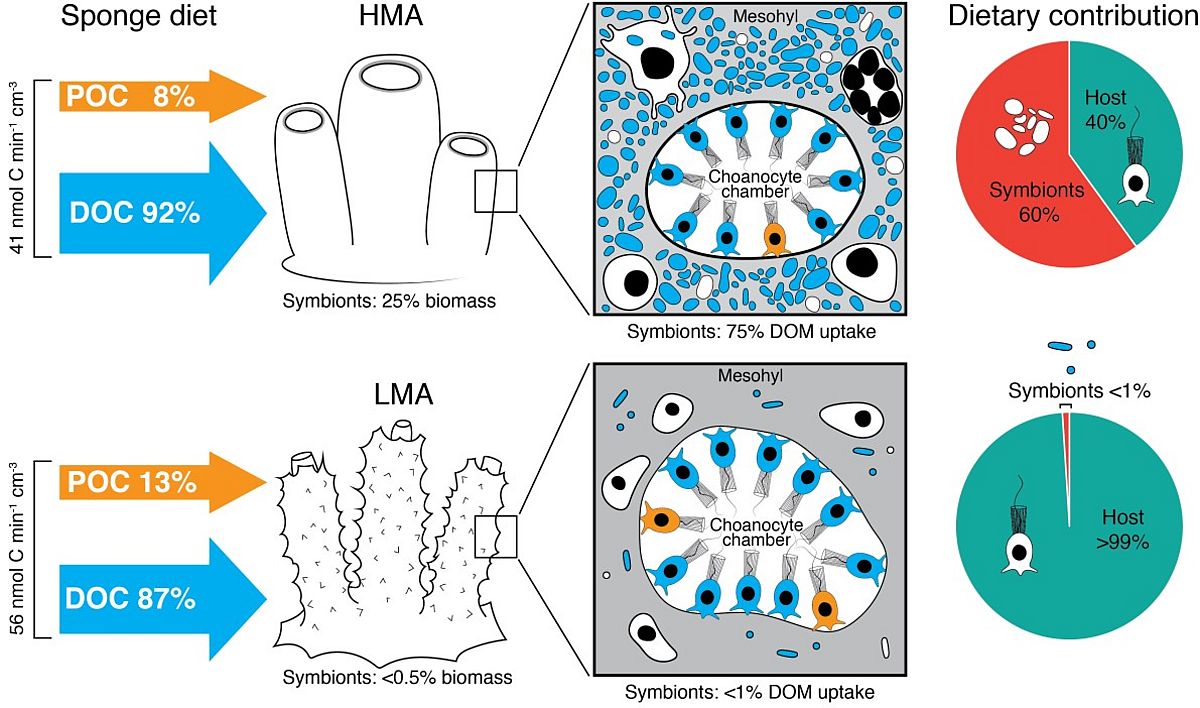
Rix L, Ribes M, Coma R, Jahn M, van Oevelen D, de Goeij J, Escrig S, Meibom A, Hentschel U (2020) Heterotrophy in the earliest gut: A single-cell view of heterotrophic carbon and nitrogen assimilation in sponge-microbe symbioses. ISME J: doi.org/10.1038/s41396-020-0706-3
Jahn MT, Arkhipova K, Markert SM, Stigloher C, Lachnit T, Pita L, Kupczok A, Ribes M, Stengel ST, Rosenstiel P, Dutilh BE, Hentschel U (2019). A phage protein aids bacterial symbionts in eukaryote immune evasion. Cell Host & Microbe 26(4):542-550.e5. doi: 10.1016/j.chom.2019.08.019.
Batani G, Bayer K, Böge J, Hentschel U, Thomas T (2019) Fluorescence in situ hybridization (FISH) and cell sorting of living bacteria. Sci Rep 9(1):18618. doi: 10.1038/s41598-019-55049-2.
Bayer K, Jahn MT, Slaby BM, Moitinho-Silva L, Hentschel U (2018) Marine sponges as Chloroflexi Hot Spots: Genomic insights and high resolution visualization of an abundant and diverse symbiotic clade. mSystems: doi.org/10.1128/mSystems.00150-18
Funding
SFB 1182/2: Origin and Function of Metaorganisms
http://www.metaorganism-research.com/